Lysozyme in water
This example will guide a new user through the usage of the @binary,
@mpi and @constraint decorators for setting up a simulation system
containing a set of proteins (lysozymes) in boxes of water with ions.
Each step contains an explanation of input and output,
using typical settings for general use.
Extracted from: http://www.mdtutorials.com/gmx/lysozyme/index.html Originally done by: Justin A. Lemkul, Ph.D. From: Virginia Tech Department of Biochemistry
Note
This example reaches up to stage 4 (energy minimization).
Important
This application requires Gromacs gmx and gmx_mpi.
from os import listdir
from os.path import isfile, join
import sys
from pycompss.api.task import task
from pycompss.api.constraint import constraint
from pycompss.api.binary import binary
from pycompss.api.mpi import mpi
from pycompss.api.parameter import *
# ############ #
# Step 1 tasks #
# ############ #
@binary(binary='${GMX_BIN}/gmx')
@task(protein=FILE_IN,
structure=FILE_OUT,
topology=FILE_OUT)
def generate_topology(mode='pdb2gmx',
protein_flag='-f', protein=None,
structure_flag='-o', structure=None,
topology_flag='-p', topology=None,
flags='-ignh',
forcefield_flag='-ff', forcefield='oplsaa',
water_flag='-water', water='spce'):
# Command: gmx pdb2gmx -f protein.pdb -o structure.gro -p topology.top -ignh -ff amber03 -water tip3p
pass
# ############ #
# Step 2 tasks #
# ############ #
@binary(binary='${GMX_BIN}/gmx')
@task(structure=FILE_IN,
structure_newbox=FILE_OUT)
def define_box(mode='editconf',
structure_flag='-f', structure=None,
structure_newbox_flag='-o', structure_newbox=None,
center_flag='-c',
distance_flag='-d', distance='1.0',
boxtype_flag='-bt', boxtype='cubic'):
# Command: gmx editconf -f structure.gro -o structure_newbox.gro -c -d 1.0 -bt cubic
pass
# ############ #
# Step 3 tasks #
# ############ #
@binary(binary='${GMX_BIN}/gmx')
@task(structure_newbox=FILE_IN,
protein_solv=FILE_OUT,
topology=FILE_IN)
def add_solvate(mode='solvate',
structure_newbox_flag='-cp', structure_newbox=None,
configuration_solvent_flag='-cs', configuration_solvent='spc216.gro',
protein_solv_flag='-o', protein_solv=None,
topology_flag='-p', topology=None):
# Command: gmx solvate -cp structure_newbox.gro -cs spc216.gro -o protein_solv.gro -p topology.top
pass
# ############ #
# Step 4 tasks #
# ############ #
@binary(binary='${GMX_BIN}/gmx')
@task(conf=FILE_IN,
protein_solv=FILE_IN,
topology=FILE_IN,
output=FILE_OUT)
def assemble_tpr(mode='grompp',
conf_flag='-f', conf=None,
protein_solv_flag='-c', protein_solv=None,
topology_flag='-p', topology=None,
output_flag='-o', output=None):
# Command: gmx grompp -f ions.mdp -c protein_solv.gro -p topology.top -o ions.tpr
pass
@binary(binary='${GMX_BIN}/gmx')
@task(ions=FILE_IN,
output=FILE_OUT,
topology=FILE_IN,
group={Type:FILE_IN, StdIOStream:STDIN})
def replace_solvent_with_ions(mode='genion',
ions_flag='-s', ions=None,
output_flag='-o', output=None,
topology_flag='-p', topology=None,
pname_flag='-pname', pname='NA',
nname_flag='-nname', nname='CL',
neutral_flag='-neutral',
group=None):
# Command: gmx genion -s ions.tpr -o 1AKI_solv_ions.gro -p topol.top -pname NA -nname CL -neutral < ../config/genion.group
pass
# ############ #
# Step 5 tasks #
# ############ #
computing_units = "24"
computing_nodes = "1"
@constraint(computing_units=computing_units)
@mpi(runner="mpirun", binary="gmx_mpi", computing_nodes=computing_nodes)
@task(em=FILE_IN,
em_energy=FILE_OUT)
def energy_minimization(mode='mdrun',
verbose_flag='-v',
ompthreads_flag='-ntomp', ompthreads='0',
em_flag='-s', em=None,
em_energy_flag='-e', em_energy=None):
# Command: gmx mdrun -v -s em.tpr
pass
# ############ #
# Step 6 tasks #
# ############ #
@binary(binary='${GMX_BIN}/gmx')
@task(em=FILE_IN,
output=FILE_OUT,
selection={Type:FILE_IN, StdIOStream:STDIN})
def energy_analisis(mode='energy',
em_flag='-f', em=None,
output_flag='-o', output=None,
selection=None):
# Command: gmx energy -f em.edr -o output.xvg
pass
# ############# #
# MAIN FUNCTION #
# ############# #
def main(dataset_path, output_path, config_path):
print("Starting demo")
protein_names = []
protein_pdbs = []
# Look for proteins in the dataset folder
for f in listdir(dataset_path):
if isfile(join(dataset_path, f)):
protein_names.append(f.split('.')[0])
protein_pdbs.append(join(dataset_path, f))
proteins = zip(protein_names, protein_pdbs)
# Iterate over the proteins and process them
result_image_paths = []
for name, pdb in proteins:
# 1st step - Generate topology
structure = join(output_path, name + '.gro')
topology = join(output_path, name + '.top')
generate_topology(protein=pdb,
structure=structure,
topology=topology)
# 2nd step - Define box
structure_newbox = join(output_path, name + '_newbox.gro')
define_box(structure=structure,
structure_newbox=structure_newbox)
# 3rd step - Add solvate
protein_solv = join(output_path, name + '_solv.gro')
add_solvate(structure_newbox=structure_newbox,
protein_solv=protein_solv,
topology=topology)
# 4th step - Add ions
# Assemble with ions.mdp
ions_conf = join(config_path, 'ions.mdp')
ions = join(output_path, name + '_ions.tpr')
assemble_tpr(conf=ions_conf,
protein_solv=protein_solv,
topology=topology,
output=ions)
protein_solv_ions = join(output_path, name + '_solv_ions.gro')
group = join(config_path, 'genion.group') # 13 = SOL
replace_solvent_with_ions(ions=ions,
output=protein_solv_ions,
topology=topology,
group=group)
# 5th step - Minimize energy
# Reasemble with minim.mdp
minim_conf = join(config_path, 'minim.mdp')
em = join(output_path, name + '_em.tpr')
assemble_tpr(conf=minim_conf,
protein_solv=protein_solv_ions,
topology=topology,
output=em)
em_energy = join(output_path, name + '_em_energy.edr')
energy_minimization(em=em,
em_energy=em_energy)
# 6th step - Energy analysis (generate xvg image)
energy_result = join(output_path, name + '_potential.xvg')
energy_selection = join(config_path, 'energy.selection') # 10 = potential
energy_analisis(em=em_energy,
output=energy_result,
selection=energy_selection)
if __name__=='__main__':
config_path = sys.argv[1]
dataset_path = sys.argv[2]
output_path = sys.argv[3]
main(dataset_path, output_path, config_path)
This application can be executed by invoking the runcompss command defining
the config_path, dataset_path and output_path where the application
inputs and outputs are. For the sake of completeness, we show how to execute
this application in a Supercomputer. In this case, the execution will be
enqueued in the supercomputer queuing system (e.g. SLURM) through the use
of the enqueue_compss command, where all parameters used in runcompss
must appear, as well as some parameters required for the queuing system (e.g. walltime).
The following code shows a bash script to submit the execution in MareNostrum IV supercomputer:
#!/bin/bash -e
# Define script variables
scriptDir=$(pwd)/$(dirname $0)
execFile=${scriptDir}/src/lysozyme_in_water.py
appClasspath=${scriptDir}/src/
appPythonpath=${scriptDir}/src/
# Retrieve arguments
numNodes=$1
executionTime=$2
tracing=$3
# Leave application args on $@
shift 3
# Load necessary modules
module purge
module load intel/2017.4 impi/2017.4 mkl/2017.4 bsc/1.0
module load COMPSs/2.7
module load gromacs/2016.4 # exposes gmx_mpi binary
export GMX_BIN=/home/user/lysozyme5.1.2/bin # exposes gmx binary
# Enqueue the application
enqueue_compss \
--num_nodes=$numNodes \
--exec_time=$executionTime \
--master_working_dir=. \
--worker_working_dir=scratch \
--tracing=$tracing \
--graph=true \
-d \
--classpath=$appClasspath \
--pythonpath=$appPythonpath \
--lang=python \
$execFile $@
######################################################
# APPLICATION EXECUTION EXAMPLE
# Call:
# ./launch_md.sh <NUMBER_OF_NODES> <EXECUTION_TIME> <TRACING> <CONFIG_PATH> <DATASET_PATH> <OUTPUT_PATH>
#
# Example:
# ./launch_md.sh 2 10 false $(pwd)/config/ $(pwd)/dataset/ $(pwd)/output/
#
#####################################################
Having the 1aki.pdb, 1u3m.pdb and 1xyw.pdb`` proteins in the dataset
folder, the execution of this script produces the submission of the job with
the following output:
$ ./launch_md.sh 2 10 false $(pwd)/config/ $(pwd)/dataset/ $(pwd)/output/
remove mkl/2017.4 (LD_LIBRARY_PATH)
remove impi/2017.4 (PATH, MANPATH, LD_LIBRARY_PATH)
Set INTEL compilers as MPI wrappers backend
load impi/2017.4 (PATH, MANPATH, LD_LIBRARY_PATH)
load mkl/2017.4 (LD_LIBRARY_PATH)
load java/8u131 (PATH, MANPATH, JAVA_HOME, JAVA_ROOT, JAVA_BINDIR, SDK_HOME, JDK_HOME, JRE_HOME)
load papi/5.5.1 (PATH, LD_LIBRARY_PATH, C_INCLUDE_PATH)
Loading default Python 2.7.13.
* For alternative Python versions, please set the COMPSS_PYTHON_VERSION environment variable with 2, 3, 2-jupyter or 3-jupyter before loading the COMPSs module.
load PYTHON/2.7.13 (PATH, MANPATH, LD_LIBRARY_PATH, LIBRARY_PATH, PKG_CONFIG_PATH, C_INCLUDE_PATH, CPLUS_INCLUDE_PATH, PYTHONHOME)
load lzo/2.10 (LD_LIBRARY_PATH,PKG_CONFIG_PATH,CFLAGS,CXXFLAGS,LDFLAGS)
load boost/1.64.0_py2 (LD_LIBRARY_PATH, LIBRARY_PATH, C_INCLUDE_PATH, CPLUS_INCLUDE_PATH, BOOST_ROOT)
load COMPSs/2.7 (PATH, CLASSPATH, MANPATH, GAT_LOCATION, COMPSS_HOME, JAVA_TOOL_OPTIONS, LDFLAGS, CPPFLAGS)
load gromacs/2016.4 (PATH, LD_LIBRARY_PATH)
SC Configuration: default.cfg
JobName: COMPSs
Queue: default
Reservation: disabled
Num Nodes: 2
Num Switches: 0
GPUs per node: 0
Job dependency: None
Exec-Time: 00:10:00
QoS: debug
Constraints: disabled
Storage Home: null
Storage Properties:
Other:
--sc_cfg=default.cfg
--qos=debug
--master_working_dir=.
--worker_working_dir=/gpfs/home/user/lysozyme
--tracing=false
--graph=true
--classpath=/home/user/lysozyme/./src/
--pythonpath=/home/user/lysozyme/./src/
--lang=python /home/user/lysozyme/./src/lysozyme_in_water.py /home/user/lysozyme/config/ /home/user/lysozyme/dataset/ /home/user/lysozyme/output/
Temp submit script is: /scratch/tmp/tmp.sMHLsaTUJj
Requesting 96 processes
Submitted batch job 10178129
Once executed, it produces the compss-10178129.out file, containing all the
standard output messages flushed during the execution:
$ cat compss-10178129.out
------ Launching COMPSs application ------
[ INFO] Using default execution type: compss
[ INFO] Relative Classpath resolved: /home/user/lysozyme/./src/:
----------------- Executing lysozyme_in_water.py --------------------------
[(590) API] - Starting COMPSs Runtime v2.7 (build 20200519-1005.r6093e5ac94d67250e097a6fad9d3ec00d676fe6c)
Starting demo
# Here it takes some time to process the dataset
[(290788) API] - Execution Finished
------------------------------------------------------------
[LAUNCH_COMPSS] Waiting for application completion
Since the execution has been performed with the task dependency graph generation enabled, the result is depicted in Figure 49. It can be identified that PyCOMPSs has been able to analyse the three given proteins in parallel.
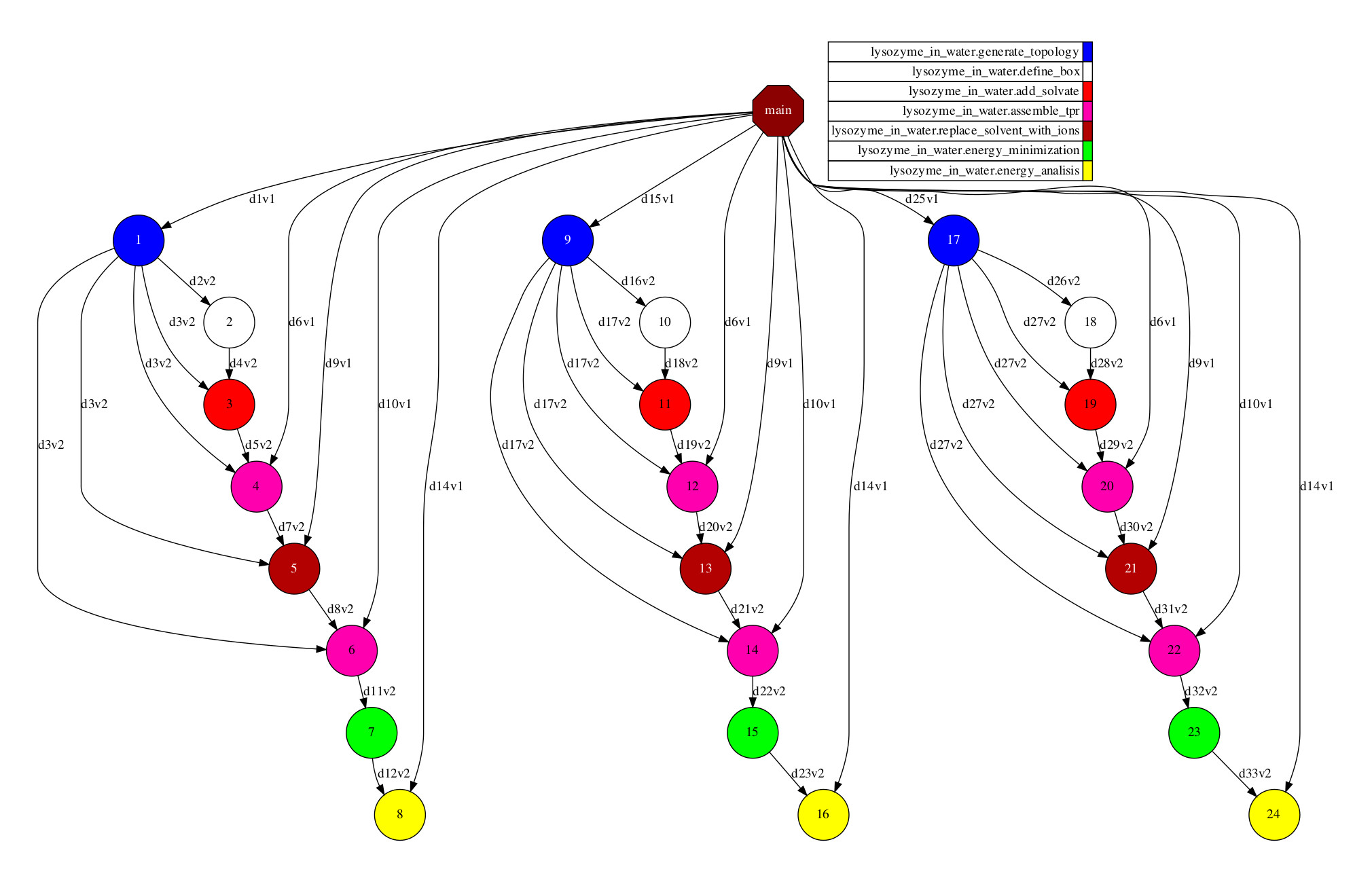
Figure 49 Python Lysozyme in Water tasks graph
The output of the application is a set of files within the output folder.
It can be seen that the files decorated with FILE_OUT are stored in this
folder. In particular, potential (.xvg) files represent the final results
of the application, which can be visualized with GRACE.
user@login:~/lysozyme/output> ls -l
total 79411
-rw-r--r-- 1 user group 8976 may 19 17:06 1aki_em_energy.edr
-rw-r--r-- 1 user group 1280044 may 19 17:03 1aki_em.tpr
-rw-r--r-- 1 user group 88246 may 19 17:03 1aki.gro
-rw-r--r-- 1 user group 1279304 may 19 17:03 1aki_ions.tpr
-rw-r--r-- 1 user group 88246 may 19 17:03 1aki_newbox.gro
-rw-r--r-- 1 user group 2141 may 19 17:06 1aki_potential.xvg <-------
-rw-r--r-- 1 user group 1525186 may 19 17:03 1aki_solv.gro
-rw-r--r-- 1 user group 1524475 may 19 17:03 1aki_solv_ions.gro
-rw-r--r-- 1 user group 577616 may 19 17:03 1aki.top
-rw-r--r-- 1 user group 577570 ene 24 16:11 #1aki.top.1#
-rw-r--r-- 1 user group 577601 may 19 16:59 #1aki.top.10#
-rw-r--r-- 1 user group 577570 may 19 17:03 #1aki.top.11#
-rw-r--r-- 1 user group 577601 may 19 17:03 #1aki.top.12#
-rw-r--r-- 1 user group 577601 ene 24 16:11 #1aki.top.2#
-rw-r--r-- 1 user group 577570 ene 24 16:20 #1aki.top.3#
-rw-r--r-- 1 user group 577601 ene 24 16:20 #1aki.top.4#
-rw-r--r-- 1 user group 577570 ene 24 16:25 #1aki.top.5#
-rw-r--r-- 1 user group 577601 ene 24 16:25 #1aki.top.6#
-rw-r--r-- 1 user group 577570 ene 24 16:31 #1aki.top.7#
-rw-r--r-- 1 user group 577601 ene 24 16:31 #1aki.top.8#
-rw-r--r-- 1 user group 577570 may 19 16:59 #1aki.top.9#
-rw-r--r-- 1 user group 8976 may 19 17:08 1u3m_em_energy.edr
-rw-r--r-- 1 user group 1416272 may 19 17:03 1u3m_em.tpr
-rw-r--r-- 1 user group 82046 may 19 17:03 1u3m.gro
-rw-r--r-- 1 user group 1415196 may 19 17:03 1u3m_ions.tpr
-rw-r--r-- 1 user group 82046 may 19 17:03 1u3m_newbox.gro
-rw-r--r-- 1 user group 2151 may 19 17:08 1u3m_potential.xvg <-------
-rw-r--r-- 1 user group 1837046 may 19 17:03 1u3m_solv.gro
-rw-r--r-- 1 user group 1836965 may 19 17:03 1u3m_solv_ions.gro
-rw-r--r-- 1 user group 537950 may 19 17:03 1u3m.top
-rw-r--r-- 1 user group 537904 ene 24 16:11 #1u3m.top.1#
-rw-r--r-- 1 user group 537935 may 19 16:59 #1u3m.top.10#
-rw-r--r-- 1 user group 537904 may 19 17:03 #1u3m.top.11#
-rw-r--r-- 1 user group 537935 may 19 17:03 #1u3m.top.12#
-rw-r--r-- 1 user group 537935 ene 24 16:11 #1u3m.top.2#
-rw-r--r-- 1 user group 537904 ene 24 16:20 #1u3m.top.3#
-rw-r--r-- 1 user group 537935 ene 24 16:20 #1u3m.top.4#
-rw-r--r-- 1 user group 537904 ene 24 16:25 #1u3m.top.5#
-rw-r--r-- 1 user group 537935 ene 24 16:25 #1u3m.top.6#
-rw-r--r-- 1 user group 537904 ene 24 16:31 #1u3m.top.7#
-rw-r--r-- 1 user group 537935 ene 24 16:31 #1u3m.top.8#
-rw-r--r-- 1 user group 537904 may 19 16:59 #1u3m.top.9#
-rw-r--r-- 1 user group 8780 may 19 17:08 1xyw_em_energy.edr
-rw-r--r-- 1 user group 1408872 may 19 17:03 1xyw_em.tpr
-rw-r--r-- 1 user group 80112 may 19 17:03 1xyw.gro
-rw-r--r-- 1 user group 1407844 may 19 17:03 1xyw_ions.tpr
-rw-r--r-- 1 user group 80112 may 19 17:03 1xyw_newbox.gro
-rw-r--r-- 1 user group 2141 may 19 17:08 1xyw_potential.xvg <-------
-rw-r--r-- 1 user group 1845237 may 19 17:03 1xyw_solv.gro
-rw-r--r-- 1 user group 1845066 may 19 17:03 1xyw_solv_ions.gro
-rw-r--r-- 1 user group 524026 may 19 17:03 1xyw.top
-rw-r--r-- 1 user group 523980 ene 24 16:11 #1xyw.top.1#
-rw-r--r-- 1 user group 524011 may 19 16:59 #1xyw.top.10#
-rw-r--r-- 1 user group 523980 may 19 17:03 #1xyw.top.11#
-rw-r--r-- 1 user group 524011 may 19 17:03 #1xyw.top.12#
-rw-r--r-- 1 user group 524011 ene 24 16:11 #1xyw.top.2#
-rw-r--r-- 1 user group 523980 ene 24 16:20 #1xyw.top.3#
-rw-r--r-- 1 user group 524011 ene 24 16:20 #1xyw.top.4#
-rw-r--r-- 1 user group 523980 ene 24 16:25 #1xyw.top.5#
-rw-r--r-- 1 user group 524011 ene 24 16:25 #1xyw.top.6#
-rw-r--r-- 1 user group 523980 ene 24 16:31 #1xyw.top.7#
-rw-r--r-- 1 user group 524011 ene 24 16:31 #1xyw.top.8#
-rw-r--r-- 1 user group 523980 may 19 16:59 #1xyw.top.9#
Figure 50 depicts the potential results obtained for the 1xyw protein.
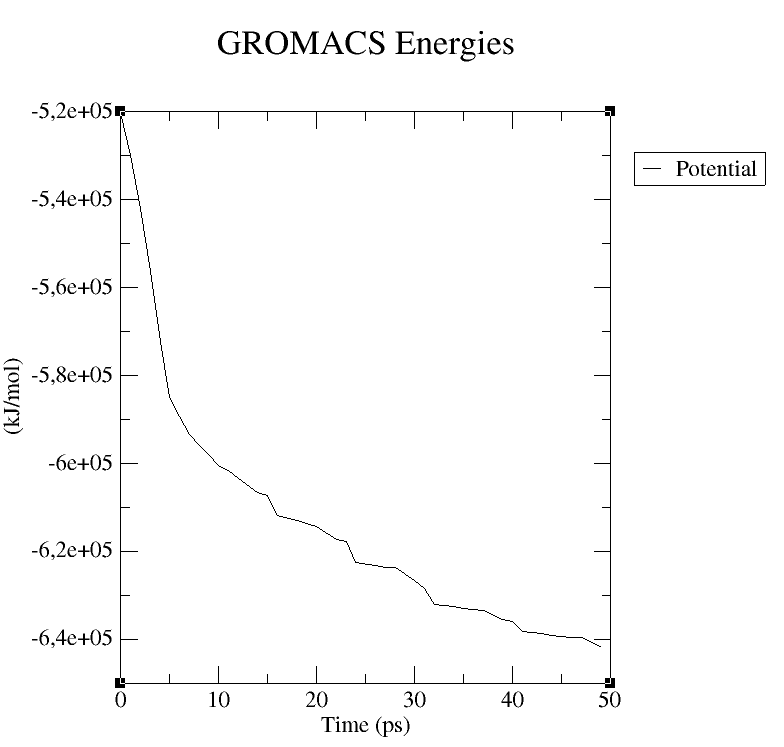
Figure 50 1xyw Potential result (plotted with GRACE)